Quasi-eigenstates
The quasi-eigenstates are defined as
If the energy \(E\) equals to an eigenvalue \(E_i\), then the quasi-eigenstate is the exact
eigenstate corresponding to \(E_i\), or a superposition of degenerate eigenstates if \(E_i\)
is degenerate. In this tutorial, we will show how to calculate the quasi-eigenstates and compare them
with the exact eigenstates. The script can be found at examples/advanced/quasi_eigen.py. We begin
with importing the necessary packages:
import numpy as np
import tbplas as tb
And define the following functions to generate primitive cell and sample with a single vacancy:
1def make_prim_cell() -> tb.PrimitiveCell:
2 """
3 Make graphene primitive cell with a single vacancy.
4
5 :return: primitive cell with vacancy
6 """
7 prim_cell = tb.make_graphene_diamond()
8
9 # Get the index of vacancy
10 dim = (17, 17, 1)
11 orb_map = tb.RegularOrbitalMap(dim, prim_cell.num_orb)
12 vac_id = orb_map.pc2sc(8, 8, 0, 0)
13
14 # Extend cell and remove orbital
15 prim_cell = tb.extend_prim_cell(prim_cell, dim=dim)
16 prim_cell.remove_orbitals([vac_id])
17 return prim_cell
18
19
20def make_sample() -> tb.Sample:
21 """
22 Make graphene sample with a single vacancy.
23
24 :return: sample with vacancy
25 """
26 prim_cell = tb.make_graphene_diamond()
27
28 # Get the index of vacancy
29 dim = (17, 17, 1)
30 orb_map = tb.RegularOrbitalMap(dim, prim_cell.num_orb)
31 vac_id = orb_map.pc2sc(8, 8, 0, 0)
32
33 # Create supercell and sample
34 super_cell = tb.SuperCell(prim_cell, dim=dim, pbc=(True, True, False),
35 vacancies=[vac_id])
36 sample = tb.Sample(super_cell)
37 return sample
We are going to create a \(17\times17\times1\) graphene sample with a vacancy in the center.
We utilize the RegularOrbitalMap class to get the index of the 0th orbital in the
\((8, 8, 0)\) cell. For the PrimitiveCell class, vacancies are added by calling
the remove_orbitals method, while for the SuperCell class they are added via the
vacancies argument.
Exact eigenstates
We define the following function to calculate the exact eigenstates:
1def wfc_diag(prim_cell: tb.PrimitiveCell) -> None:
2 """
3 Calculate wave function using exact diagonalization.
4
5 :param prim_cell: primitive cell
6 :return: None
7 """
8 solver = tb.DiagSolver(prim_cell)
9 solver.config.prefix = "diag"
10 solver.config.k_points = np.array([[0.0, 0.0, 0.0]])
11 bands, states = solver.calc_states()
12 i_b = prim_cell.num_orb // 2
13 wfc = np.abs(states[0, i_b])**2
14 wfc /= wfc.max()
15 vis = tb.Visualizer()
16 vis.plot_wfc(prim_cell, wfc, scatter=True, site_size=wfc * 100, site_color="r",
17 with_model=True, model_style={"alpha": 0.25, "color": "gray"},
18 fig_name="diag.png", fig_size=(6, 4.5), fig_dpi=100)
In line 8 we create the solver from DiagSolver class. Then we define the prefix of data files
and the k-points on which the eigenstates will be calculated in line 9-10, where we consider the
\(\Gamma\) point. In line 11 we call the calc_states method to get the eigenvalues and eigenstates,
as returned in bands and states. After that, we extract the eigenstate at the Fermi level and
normalize it in line 12-14. To visualize the eigenstate, we create a visualizer from the Visualizer
class and call its plot_wfc method. Note that we must use scatter plot by setting the scatter argument
to true. The eigenstate should be passed as the site_size argument, i.e., sizes of scatters will indicate the projection of eigenstate
on the sites. We also need to show the model alongside the eigenstate through the with_model argument
and define its plotting style with the model_style argument.
Quasi-eigenstates
The quasi-eigenstate is evaluated and plotted in a similar approach:
1def wfc_tbpm(sample: tb.Sample) -> None:
2 """
3 Calculate wave function using TBPM.
4
5 :param sample: graphene sample
6 :return: None
7 """
8 solver = tb.TBPMSolver(sample)
9 solver.config.prefix = "tbpm"
10 solver.config.rescale = 9.0
11 solver.config.num_random_samples = 1
12 solver.config.num_time_steps = 1024
13 solver.config.qe_energies = np.array([0.0])
14 qs = solver.calc_quasi_eigenstates()
15 wfc = qs[0] # qs is already the squared norm
16 wfc /= wfc.max()
17 vis = tb.Visualizer()
18 vis.plot_wfc(sample, wfc, scatter=True, site_size=wfc*100, site_color="b",
19 with_model=True, model_style={"alpha": 0.25, "color": "gray"},
20 fig_name="tbpm.png", fig_size=(6, 4.5), fig_dpi=100)
Evaluating quasi-eigenstates is a kind of TBPM calculation, so it follows the common procedure of
TBPM. We firstly create the solver from TBPMSolver class in line 8, then set the calculation
parameters in line 9-13. Especially, the qe_energies argument defines the energies for which the
quasi-eigenstates will be evaluated. Then we call the calc_quasi_eigenstates method of
TBPMSolver to get the eigenstates, normalize it, and visualize it in line 14-20.
Finally, we define the driving function:
1def main():
2 wfc_diag(make_prim_cell())
3 wfc_tbpm(make_sample())
4
5
6if __name__ == "__main__":
7 main()
The output is shown in Fig. 1. It is clear that the eigenstate and quasi-eigenstate are localized around the vacancy with a 3-fold rotational symmetry.
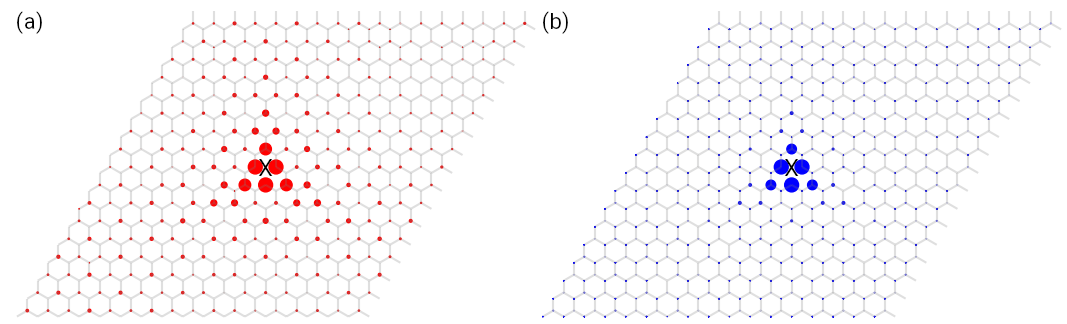
Spatial distribution of (a) exact eigenstate and (b) quasi-eigenstate of graphene sample with a vacancy in the
center. The X marks indicate the position of the vacancy.