Z2 topological invariant
In this tutorial, we demonstrate the usage of Z2 class by calculating the \(\mathbb{Z}_2\)
topological invariant of bilayer bismuth.
The corresponding script is located at examples/prim_cell/z2/bismuth.py. We begin with importing
the necessary packages:
from math import sqrt, pi
import numpy as np
from numpy.linalg import norm
import tbplas as tb
Auxiliary functions
We define the following functions to build bilayer bismuth with SOC:
1def calc_hop(sk: tb.SK, rij: np.ndarray, label_i: str, label_j: str) -> complex:
2 """
3 Evaluate the hopping integral <i,0|H|j,r>.
4
5 References:
6 [1] https://journals.aps.org/prb/abstract/10.1103/PhysRevB.84.075119
7 [2] https://journals.aps.org/prb/abstract/10.1103/PhysRevB.52.1566
8 [3] https://journals.aps.org/prl/abstract/10.1103/PhysRevLett.97.236805
9
10 :param sk: SK instance
11 :param rij: displacement vector from orbital i to j in nm
12 :param label_i: label of orbital i
13 :param label_j: label of orbital j
14 :return: hopping integral in eV
15 """
16 # Range-dependent slater-Koster parameters from ref. 2
17 dict1 = {"v_sss": -0.608, "v_sps": 1.320, "v_pps": 1.854, "v_ppp": -0.600}
18 dict2 = {"v_sss": -0.384, "v_sps": 0.433, "v_pps": 1.396, "v_ppp": -0.344}
19 dict3 = {"v_sss": 0.0, "v_sps": 0.0, "v_pps": 0.156, "v_ppp": 0.0}
20 r_norm = norm(rij)
21 if abs(r_norm - 0.30628728) < 1.0e-5:
22 data = dict1
23 elif abs(r_norm - 0.35116131) < 1.0e-5:
24 data = dict2
25 else:
26 data = dict3
27 lm_i = label_i.split(":")[1]
28 lm_j = label_j.split(":")[1]
29 return sk.eval(r=rij, label_i=lm_i, label_j=lm_j,
30 v_sss=data["v_sss"], v_sps=data["v_sps"],
31 v_pps=data["v_pps"], v_ppp=data["v_ppp"])
32
33
34def make_cell() -> tb.PrimitiveCell:
35 """
36 Make bilayer bismuth primitive cell without SOC.
37
38 :return: bilayer bismuth primitive cell
39 """
40 # Lattice constants from ref. 2
41 a = 4.5332
42 c = 11.7967
43 mu = 0.2341
44
45 # Lattice vectors of bulk from ref. 2
46 a1 = np.array([-0.5*a, -sqrt(3)/6*a, c/3])
47 a2 = np.array([0.5*a, -sqrt(3)/6*a, c/3])
48 a3 = np.array([0, sqrt(3)/3*a, c/3])
49
50 # Lattice vectors and atomic positions of bilayer from ref. 2 & 3
51 a1_2d = a2 - a1
52 a2_2d = a3 - a1
53 a3_2d = np.array([0, 0, c])
54 lat_vec = np.array([a1_2d, a2_2d, a3_2d])
55 atom_position = np.array([[0, 0, 0], [1/3, 1/3, 2*mu-1/3]])
56
57 # Create cell and add orbitals with energies from ref. 2
58 cell = tb.PrimitiveCell(lat_vec, unit=tb.ANG)
59 atom_label = ("Bi1", "Bi2")
60 e_s, e_p = -10.906, -0.486
61 orbital_energy = {"s": e_s, "px": e_p, "py": e_p, "pz": e_p}
62 for i, pos in enumerate(atom_position):
63 for orbital, energy in orbital_energy.items():
64 label = f"{atom_label[i]}:{orbital}"
65 cell.add_orbital(pos, label=label, energy=energy)
66
67 # Add hopping terms
68 neighbors = tb.find_neighbors(cell, a_max=5, b_max=5, max_distance=0.454)
69 sk = tb.SK()
70 for term in neighbors:
71 i, j = term.pair
72 label_i = cell.get_orbital(i).label
73 label_j = cell.get_orbital(j).label
74 hop = calc_hop(sk, term.rij, label_i, label_j)
75 cell.add_hopping(term.rn, i, j, hop)
76 return cell
77
78
79def add_soc(cell: tb.PrimitiveCell) -> tb.PrimitiveCell:
80 """
81 Add spin-orbital coupling to the primitive cell.
82
83 :param cell: primitive cell to modify
84 :return: primitive cell with soc
85 """
86 # Double the orbitals and hopping terms
87 cell = tb.merge_prim_cell(cell, cell)
88
89 # Add spin notations to the orbitals
90 num_orb_half = cell.num_orb // 2
91 num_orb_total = cell.num_orb
92 for i in range(num_orb_half):
93 label = cell.get_orbital(i).label
94 cell.set_orbital(i, label=f"{label}:up")
95 for i in range(num_orb_half, num_orb_total):
96 label = cell.get_orbital(i).label
97 cell.set_orbital(i, label=f"{label}:down")
98
99 # Add SOC terms
100 soc_lambda = 1.5 # ref. 2
101 soc = tb.SOC()
102 for i in range(num_orb_total):
103 label_i = cell.get_orbital(i).label.split(":")
104 atom_i, lm_i, spin_i = label_i
105
106 for j in range(i+1, num_orb_total):
107 label_j = cell.get_orbital(j).label.split(":")
108 atom_j, lm_j, spin_j = label_j
109
110 if atom_j == atom_i:
111 soc_intensity = soc.eval(label_i=lm_i, spin_i=spin_i,
112 label_j=lm_j, spin_j=spin_j)
113 soc_intensity *= soc_lambda
114 if abs(soc_intensity) >= 1.0e-15:
115 try:
116 energy = cell.get_hopping((0, 0, 0), i, j)
117 except tb.PCHopNotFoundError:
118 energy = 0.0
119 energy += soc_intensity
120 cell.add_hopping((0, 0, 0), i, j, energy)
121 return cell
Most of them are similar to that of Slater-Koster formulation and Spin-orbital coupling.
Evaluation of Z2
With all the auxiliary functions ready, we now proceed to calculate the \(\mathbb{Z}_2\) invariant number of bilayer bismuth as
1def main():
2 # Create cell and add soc
3 cell = make_cell()
4 cell = add_soc(cell)
5
6 # Evaluate Z2
7 z2 = tb.Z2(cell)
8 z2.config.prefix = "bi"
9 z2.config.num_occ = 10
10 z2.config.ka_array = np.linspace(-0.5, 0.5, 200)
11 z2.config.kb_array = np.linspace(0.0, 0.5, 200)
12 z2.config.kc = 0.0
13 kb_array, phases = z2.calc_phases()
14
15 # Plot phases
16 vis = tb.Visualizer()
17 vis.plot_phases(kb_array, phases / pi)
18
19
20if __name__ == "__main__":
21 main()
To calculate \(\mathbb{Z}_2`\) we need to create a Z2 instance. Bilayer bismuth has
two Bi atoms, each carrying two \(6s\) and three \(6p\) electrons, totally 10 electrons per
primitive cell. So the number of occupied bands is thus 10, as specified by the num_occ argument.
Then we sample \(\mathbf{k}_a\) from \(-\frac{1}{2}\mathbf{G}_a\) to \(\frac{1}{2}\mathbf{G}_a\),
and \(\mathbf{k}_b\) from \(\mathbf{0}\) to \(\frac{1}{2}\mathbf{G}_b\) via the ka_array
and kb_array arguments. kc is set to 0 since the model is two-dimensional. After that, we call
the calc_phases function to get the topological phases \(\theta_m^D\), and \(\theta_m^D\) as the
function of \(\mathbf{k}_b\) in the left panel of the figure. It is clear that the crossing
number of phases against the reference line is 1, indicating that bilayer bismuth is a topological
insulator. We then decrease the SOC intensity \(\lambda`\) to 0.15 eV and re-calculate the
phases. The results are shown in the right panel of the figure, where the crossing number is 0,
indicating that bilayer bismuth becomes a normal insulator under weak SOC, similar to the case of
bilayer Sb.
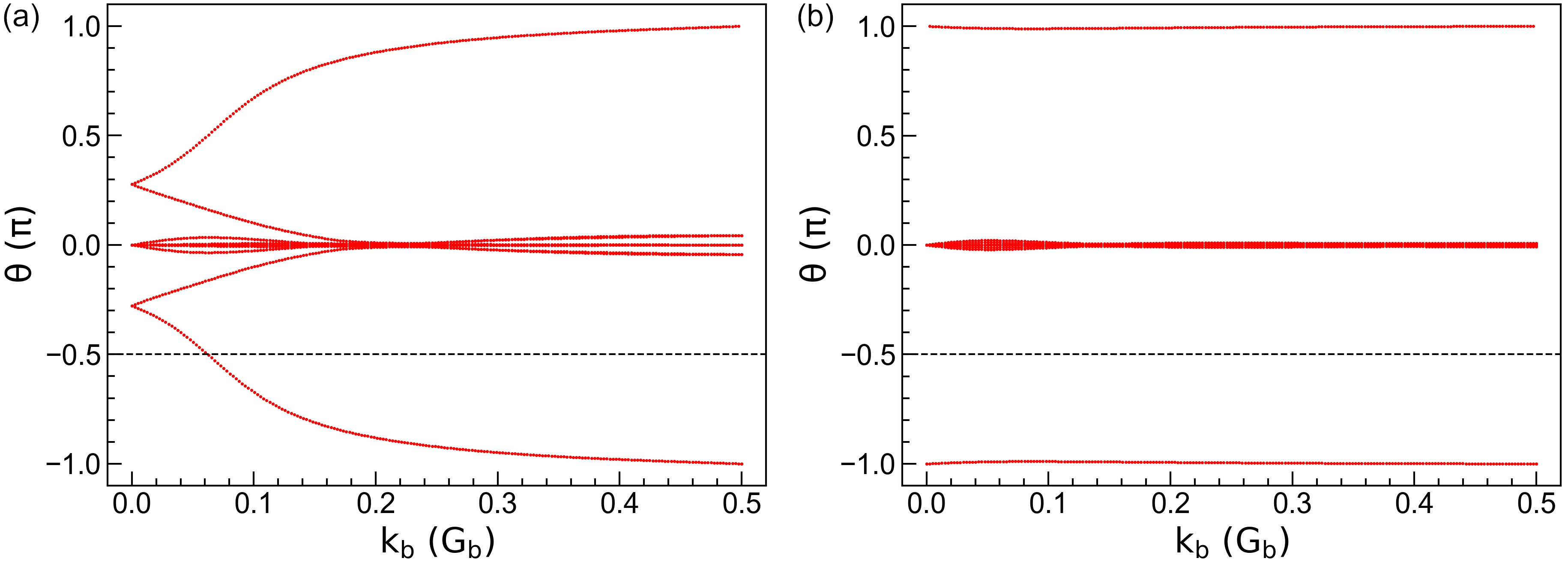
Topological phases \(\theta_m^D\) of bilayer bismuth under SOC intensity of (a) \(\lambda\) = 1.5 eV and (b) \(\lambda\) = 0.15 eV. The horizontal dashed lines indicates the reference lines with which the crossing number is determined.