Build hetero-structure
In this tutorial, we show how to build hetero-structure taking twisted bilayer graphene as the
example. The workflow of constructing hetero structures is shown in Fig. 1(a). First of all, we
determine the twisting angle and lattice vectors of the hetero-structure. Then we build the
primitive cells of each layer, shift the twisted layer along \(z\)-axis by the interlayer
distance and rotate it by the twisting angle. After that, we reshape the primitive cells to the
lattice vectors of the hetero-structure to yield the layers, as depicted in Fig. 1(b). When all the
layers are ready, we merge them into one cell and add the intralayer and interlayer hopping terms
up to a given cutoff distance. For the visualization of Moiré pattern, we also need to build a
sample from the merged cell. The script can be found at examples/advanced/tbg.py. Before
constructing the model, we need to import the required packages:
import math
from typing import Tuple, Union
import numpy as np
from numpy.linalg import norm
import tbplas as tb
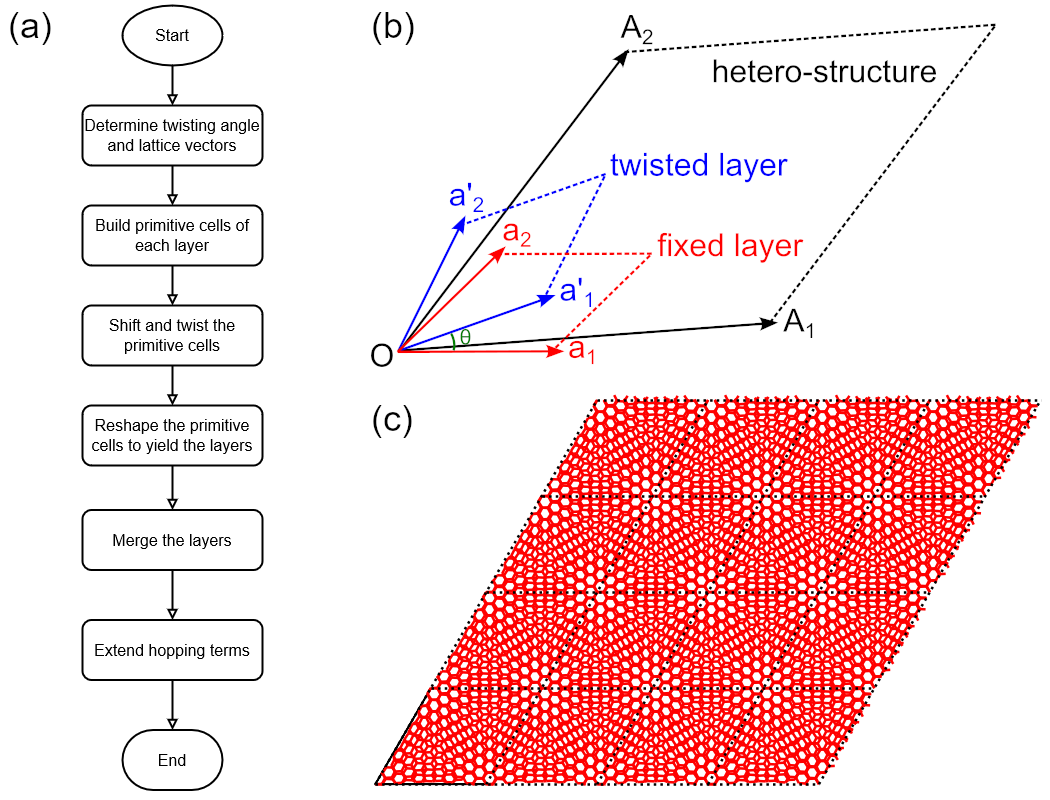
(a) Workflow of constructing hetero-structure. (b) Schematic plot of lattice vectors of fixed (\(\mathbf{a}_1\), \(\mathbf{a}_2\)) and twisted (\(\mathbf{a}_1^\prime\), \(\mathbf{a}_2^\prime\)) primitive cells and the hetero-structure (\(\mathbf{A}_1\), \(\mathbf{A}_2\)), as well as the twisting angle \(\theta\). (c) Twisted bilayer graphene sample with \(4\times4\times1\) merged cells of \(i=5\).
Auxiliary functions
The twisting angle and lattice vectors are determined following the formulation in ref1:
where \(\mathbf{a}_1\) and \(\mathbf{a}_2\) are the lattice vectors of the primitive cell of fixed layer and \(i\) is the index of hetero-structure. We define the following functions accordingly:
1def calc_twist_angle(i: int) -> float:
2 """
3 Calculate twisting angle according to ref. [1].
4
5 :param i: parameter controlling the twisting angle
6 :return: twisting angle in RADIANs, NOT degrees
7 """
8 cos_ang = (3 * i ** 2 + 3 * i + 0.5) / (3 * i ** 2 + 3 * i + 1)
9 return math.acos(cos_ang)
10
11
12def calc_hetero_lattice(i: int, prim_cell_fixed: tb.PrimitiveCell) -> np.ndarray:
13 """
14 Calculate Cartesian coordinates of lattice vectors of hetero-structure
15 according to ref. [1].
16
17 :param i: parameter controlling the twisting angle
18 :param prim_cell_fixed: primitive cell of fixed layer
19 :return: (3, 3) float64 array, Cartesian coordinates of hetero-structure
20 lattice vectors in NANOMETER
21 """
22 hetero_lattice = np.array([[i, i + 1, 0],
23 [-(i + 1), 2 * i + 1, 0],
24 [0, 0, 1]])
25 hetero_lattice = tb.frac2cart(prim_cell_fixed.lat_vec, hetero_lattice)
26 return hetero_lattice
calc_twist_angle returns the twisting angle in radians, while calc_hetero_lattice returns
the Cartesian coordinates of lattice vectors in nm. After merging the layers, we need to add the
interlayer hopping terms. Meanwhile, the intralayer hopping terms should also be extended in the
same approach. We define the extend_hop function to achieve these goals:
1def extend_hop(model: Union[tb.PrimitiveCell, tb.SuperCell],
2 max_distance: float = 0.75) -> None:
3 """
4 Extend the hopping terms in model up to cutoff distance.
5
6 :param model: model to extend
7 :param max_distance: cutoff distance in NM
8 :return: None. Incoming model is modified
9 """
10 neighbors = tb.find_neighbors(model, a_max=1, b_max=1,
11 max_distance=max_distance)
12 for term in neighbors:
13 i, j = term.pair
14 model.add_hopping(term.rn, i, j, calc_hop(term.rij))
Here we call the find_neighbors() function to get the neighboring orbital pairs up to the cutoff
distance max_distance. Then the hopping terms are evaluated according to the displacement
vector rij with the calc_hop function and added to the primitive cell. The calc_hop
function is defined according to the formulation in
ref2:
1def calc_hop(rij: np.ndarray) -> float:
2 """
3 Calculate hopping parameter according to Slater-Koster relation.
4 See ref. [2] for the formulae.
5
6 :param rij: (3,) array, displacement vector between two orbitals in NM
7 :return: hopping parameter in eV
8 """
9 a0 = 0.1418
10 a1 = 0.3349
11 r_c = 0.6140
12 l_c = 0.0265
13 gamma0 = 2.7
14 gamma1 = 0.48
15 decay = 22.18
16 q_pi = decay * a0
17 q_sigma = decay * a1
18 dr = norm(rij).item()
19 n = rij.item(2) / dr
20 v_pp_pi = - gamma0 * math.exp(q_pi * (1 - dr / a0))
21 v_pp_sigma = gamma1 * math.exp(q_sigma * (1 - dr / a1))
22 fc = 1 / (1 + math.exp((dr - r_c) / l_c))
23 hop = (n**2 * v_pp_sigma + (1 - n**2) * v_pp_pi) * fc
24 return hop
Build the model
With all the functions ready, we proceed to build the hetero-structure. Firstly, we evaluate the
twisting angle of bilayer graphene for \(i=5\). Then we construct the primitive cells of the
fixed and twisted layers with the make_graphene_diamond() function. The fixed primitive cell
is located at \(z=0\) and does not need rotation or shifting. On the other hand, the twisted
primitive cell needs to be rotated counter-clockwise by the twisting angle and shifted towards
\(+z\) by 0.3349 nm, which is done with the spiral_prim_cell() function. After that, we
reshape the primitive cells to the lattice vectors of hetero-structure with the
make_hetero_layer() function, which is a wrapper to coordinate conversion and
reshape_prim_cell(). Then the layers are merged with merge_prim_cell() and the hopping
terms are extended with extend_hop using a cutoff distance of 0.75 nm. Finally, a sample with
\(4\times4\times1\) merged cells is created and plotted, with the hopping terms below 0.3 eV
hidden for clarity. The output is shown in Fig. 1(c), where the Moiré pattern can be clearly
observed.
1def main():
2 # Evaluate twisting angle
3 i = 5
4 angle = calc_twist_angle(i)
5
6 # Prepare primitive cells of fixed and twisted layer
7 prim_cell_fixed = tb.make_graphene_diamond()
8 prim_cell_twisted = tb.make_graphene_diamond()
9
10 # Shift and rotate the twisted layer
11 tb.spiral_prim_cell(prim_cell_twisted, angle=angle, shift=0.3349)
12
13 # Reshape primitive cells to the lattice vectors of hetero-structure
14 hetero_lattice = calc_hetero_lattice(i, prim_cell_fixed)
15 layer_fixed = tb.make_hetero_layer(prim_cell_fixed, hetero_lattice)
16 layer_twisted = tb.make_hetero_layer(prim_cell_twisted, hetero_lattice)
17
18 # Merge layers
19 merged_cell = tb.merge_prim_cell(layer_fixed, layer_twisted)
20
21 # Extend hopping terms
22 extend_hop(merged_cell, max_distance=0.75)
23
24 # Visualize Moire pattern
25 sample = tb.Sample(tb.SuperCell(merged_cell, dim=(4, 4, 1), pbc=(True, True, False)))
26 sample.plot(with_orbitals=False, hop_as_arrows=False, hop_eng_cutoff=0.3)
27
28
29if __name__ == "__main__":
30 main()